Research
Gene expression levels are precisely regulated during development; quantitative perturbations to this process are a frequent cause of both rare and common human disease. However, most experimental tools that have been applied to study gene function are binary in nature. Therefore, our overarching goal is to develop and apply tools to quantitatively perturb gene regulation to understand both basic mechanisms of gene regulation and how this goes awry in disease.
Transcription factors (TFs), proteins that bind to noncoding regulatory DNA such as enhancers and promoters and modulate the production of RNA from target genes, are key drivers of developmental regulatory programs. TFs are highly dosage-sensitive; quantitative changes of 50% or less in TF levels frequently cause rare disorders or common trait variation.
We recently developed an approach to precisely modulate TF dosage in human stem cells and their derivatives; we are now using such tools to build quantitative, predictive models of how TF dosage impacts molecular, cellular, and morphological phenotypes, with the goal of identifying the regulatory elements, genes, and cell states that are most sensitive to TF dosage and thus drivers of disease.
Areas of focus
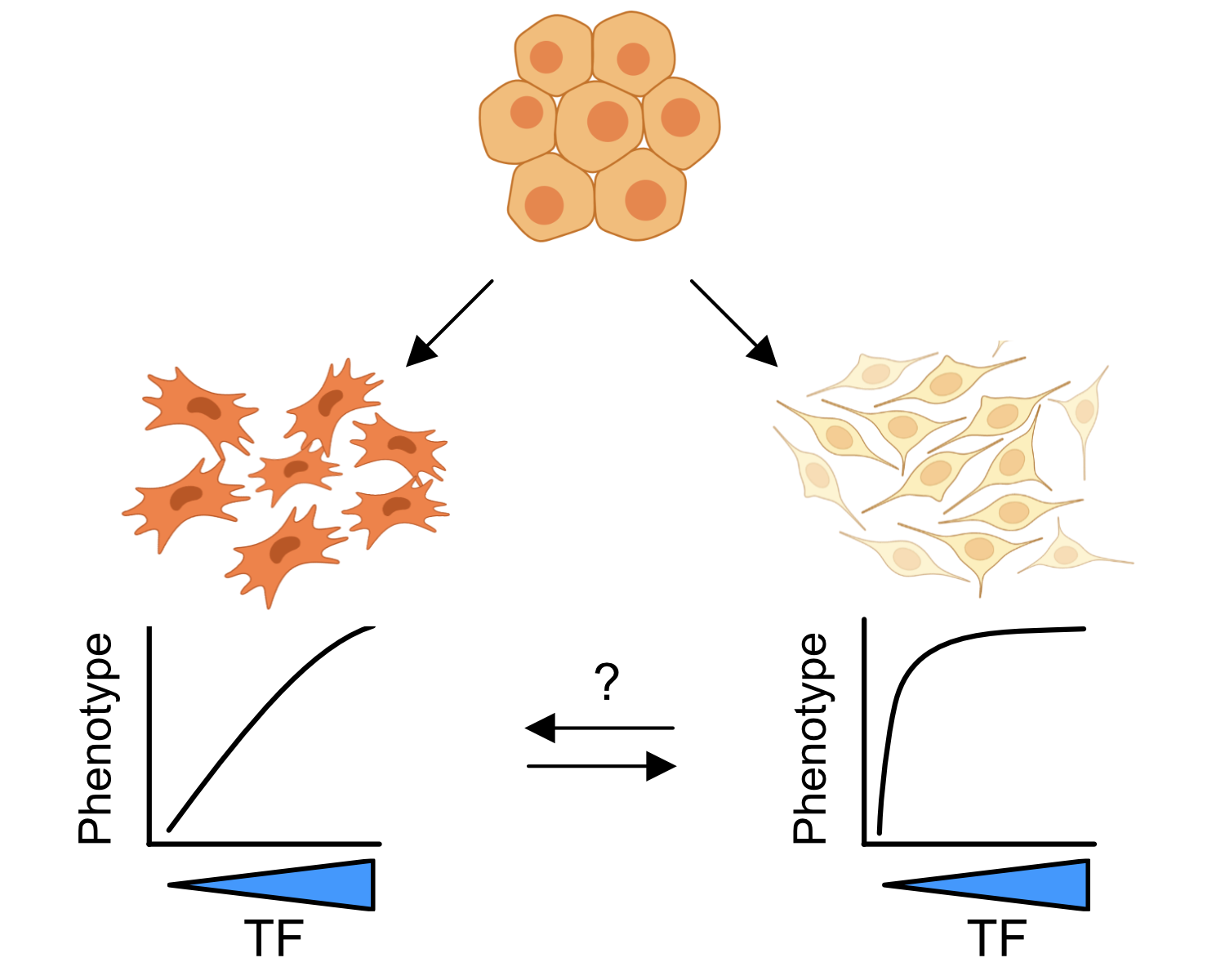
Cell type specificity of TF dosage sensitivity
In addition to being dosage-sensitive, TFs are pleiotropic - one TF can have important roles in multiple cell types. Despite these broad roles, TF dosage sensitivity typically impacts one or a few cell types. To understand what drives this specificity, we study the human neural crest, a transient, embryonic cell population that gives rise to a fascinating array of cell types and tissues. Understanding why specific neural crest lineages are sensitive to dosage of broadly important TFs will yield new insights into developmental disorders that affect organ systems as diverse as the face, as in craniofacial malformations, and the gut, such as the loss of the enteric nervous system in Hirschsprung’s disease.
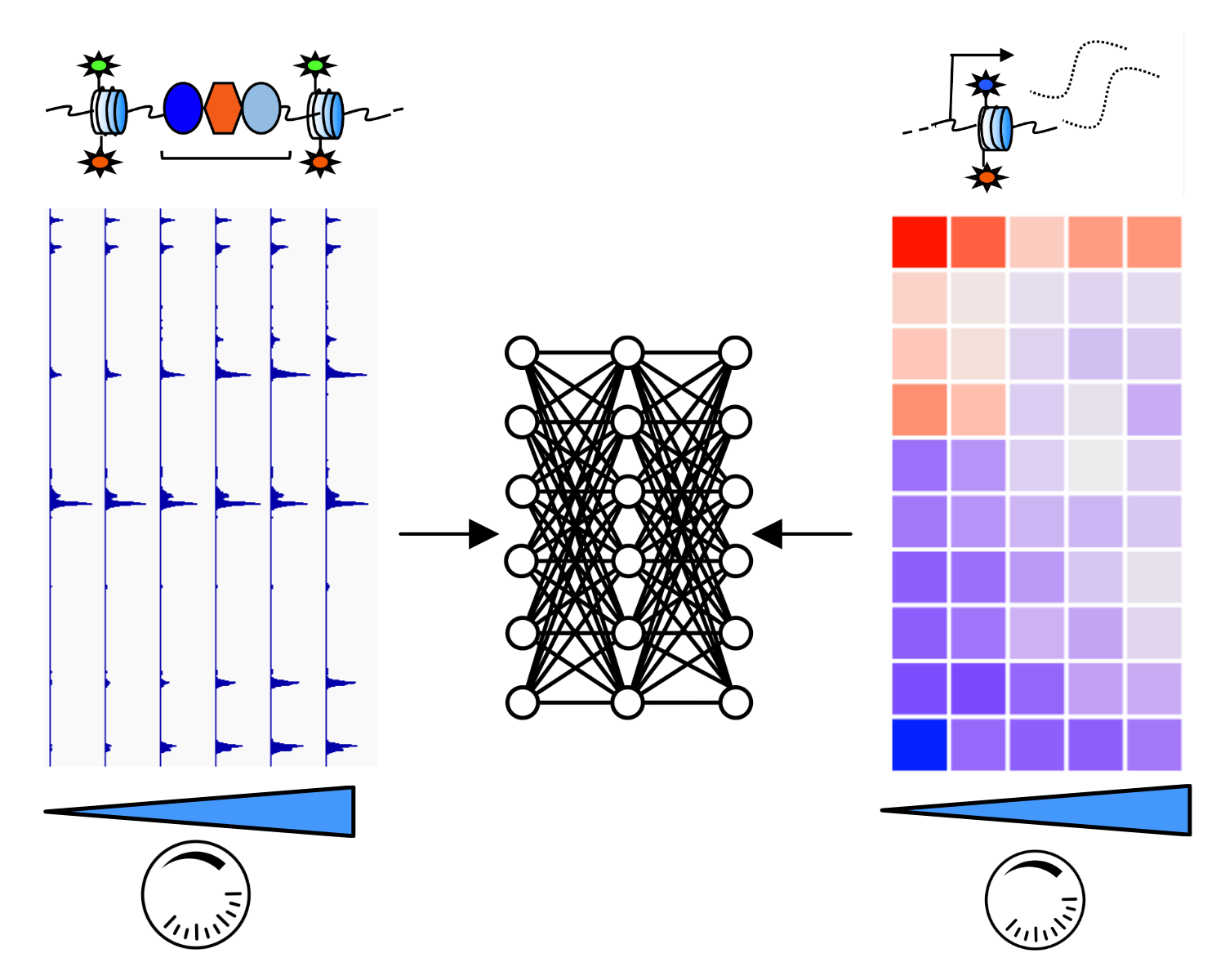
Machine learning to decode gene regulatory mechanisms
Recent advances in machine learning and artificial intelligence have made substantial advances in learning the sequence features that predict cell type-specific chromatin and gene regulatory networks. However, the vast majority of these approaches rely on static, steady-state measurements. We are combining deep learning approaches with quantitative perturbations and functional genomic data to reveal hidden layers of the cis-regulatory code that predict responsiveness of enhancers and genes to TF dosage. Ultimately, we aim to accurately predict quantitative TF dosage effects in any cell type, from sequence alone.
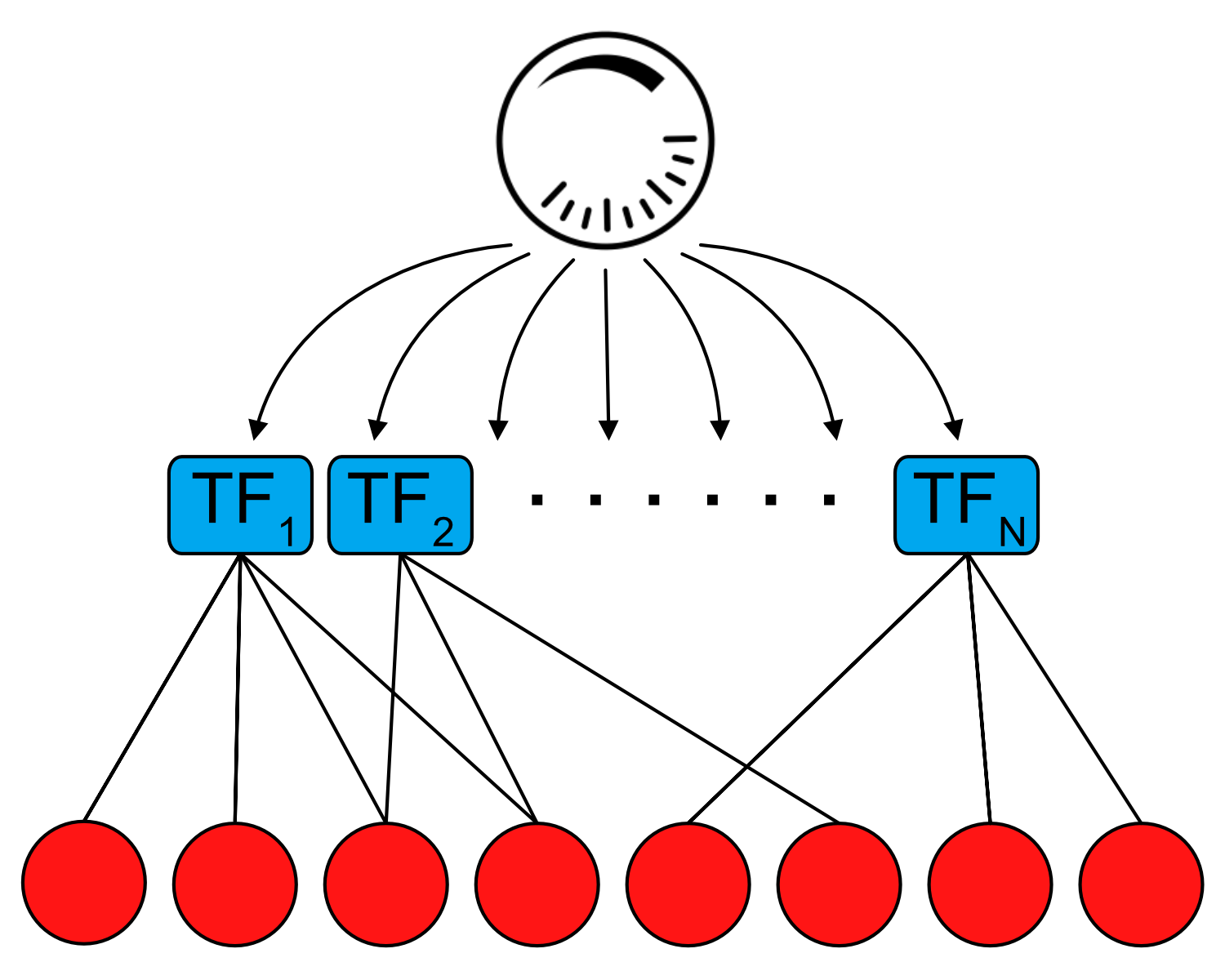
Expanding the scope and scale of quantitative perturbations
We and others have developed and applied chemical biology-based approaches that can precisely manipulate protein levels, but with low throughput. CRISPR-based methods for titrating gene dosage are higher throughput, but with lower precision. We aim to develop novel techniques for precise manipulation of protein levels for dozens to hundreds of genes in a single experiment. Such methods could be applied to essentially every expressed TF in a given cell type, allowing us to query why some TFs are more dosage-sensitive than others and provide a quantitative view of gene regulatory networks otherwise mapped through binary perturbations.
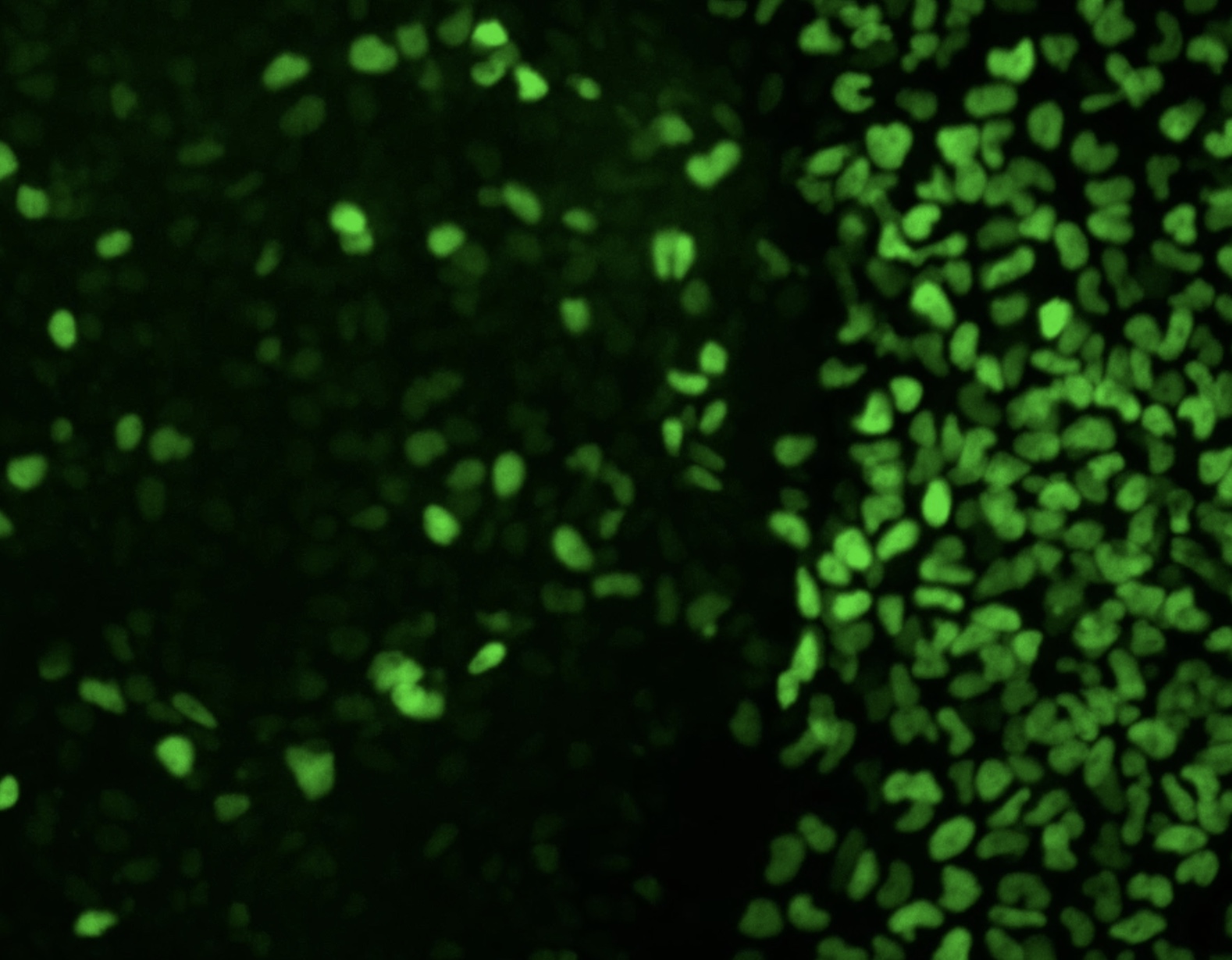
From TF dosage to cellular and morphological outcomes
While TFs are defined by their molecular function as regulators of gene expression, their dosage changes ultimately cause disease by resulting in specific changes to cellular behavior and/or tissue morphology and growth. We are combining stem cell-derived organoid models of human development with functional genomics live-cell imaging to understand how quantitative changes in TF dosage percolate through different levels of scale to ultimately result in tissue-level phenotypes. This integrative approach could reveal novel therapeutic approaches to prevent or reverse dosage-sensitive developmental disorders.